Cell adhesion is critical for the survival of most cells. Adhesion factors influence cell behavior, such as migration and proliferation. Traditional high-throughput screenings for adhesion factors are resource-intensive. Sonnentag et al. introduced high-density peptide arrays to identify peptides that influence cell adhesion or detachment. They used a large library of unique fragments and analyzed cells on multifunctionalized surfaces. The study identified cell-repellent and adhesive peptides, demonstrating the efficiency of the method in discovering functional peptides that modulate cell behavior.
Background
Adhesion to a substrate is a survival requirement for most cells. In normal tissues, cells attach to the basement membrane of the extracellular matrix (ECM). The ECM is composed of macromolecules that collectively influence cell behavior by regulating migration, differentiation, survival, and proliferation. Therefore, understanding which factors display adhesive and repellent properties is key for several biomedical fields, such as tissue engineering, drug delivery, and the development of biomaterials.
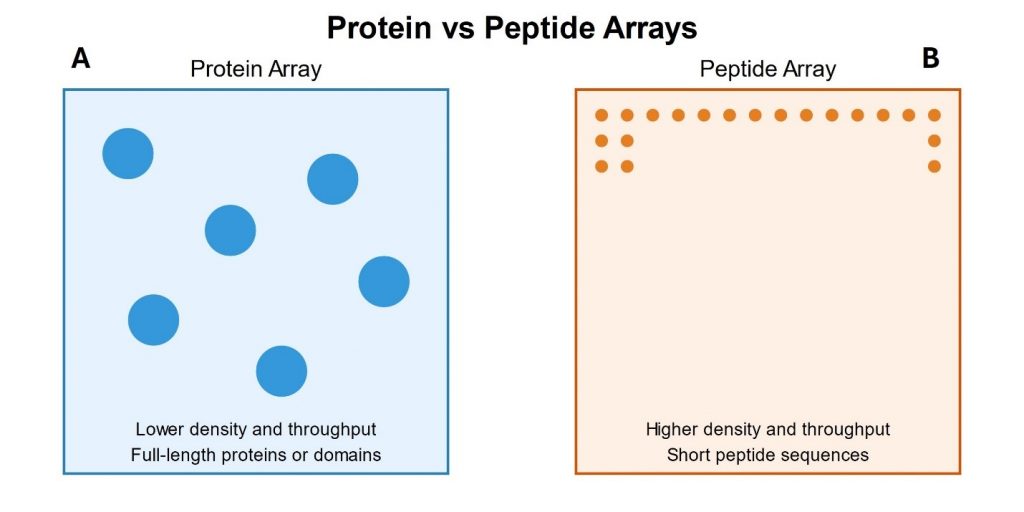
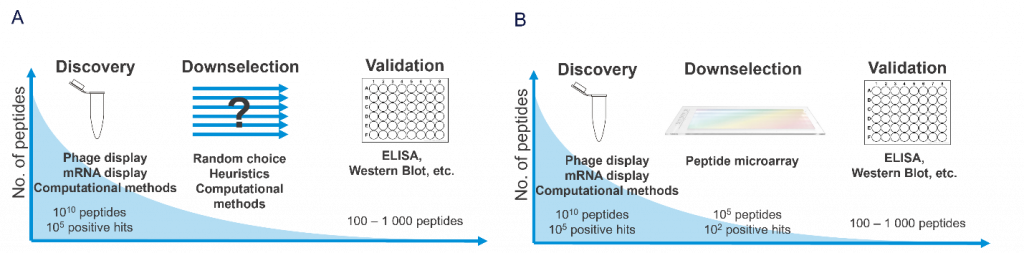
High-throughput screening (HTS) and miniaturized platforms have been used to systematically identify factors affecting cell adhesion. However, these traditional methods are often highly resource-consuming and have considerable technical limitations. Sonnentag et al. [1] described the use of high-density peptide arrays as a new tool to screen for extracellular factors that promote the adhesion or detachment of cancer cells on multifunctionalized surfaces.
Methodology
Firstly, the authors designed a peptide library containing 11,314 unique fragments for three groups of peptides:
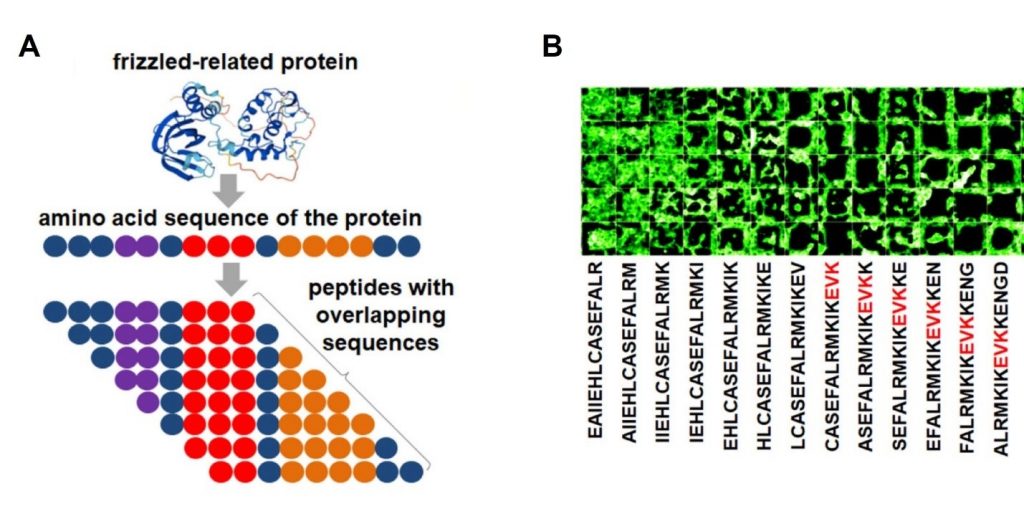
- Secreted frizzled-related protein (SFRP1) (Figure 1), Dickkopf-related protein (DKK1), nd tumor necrosis factor (TNF).
- 726 substitution sequences derived from four peptides reported to inhibit the co-receptor function of CD44v6 for the receptor tyrosine kinase MET and thus inhibit cancer cell growth and metastasis.
- Sequences based on random combinations of peptide fragments from the first group.
Secondly, the authors transduced SW620 mCherry colorectal cancer cells with the TOP-GFP construct, a Wnt-pathway reporter driving the expression of green fluorescent protein.
Thirdly, the authors transferred the cells onto the chip and analyzed them by confocal fluorescence microscopy.
Key findings
By designing the arrays with peptides overlapping by one amino acid, and spanning the entire protein sequence (Figure 1), the authors were able to conclude the following:
- The EVK motif is a strong repellent for cells (Figure 1B).
- Fragments from the DKK1 and SFRP1 proteins, as well as a sequence with a single mutation in the CD44v6-inhibiting peptide, have strong cell-repellent capacity. The authors further mapped the identified peptides to the 3D structures of the corresponding proteins and found them not to be associated with specific secondary structures.
- Peptides with strong adhesive properties are short and similar to other known protein sequences. This suggests that cell-peptide interactions are highly selective and protein-protein binding occurs due to the recognition of short domains by more complex binding grooves.
- Cell migration occurs from repulsive to adhesive regions after an initial random cell distribution over a surface. The authors demonstrated this by placing the cells on a surface with alternating patterns of cell-adhesive and cell-repellent peptides.
Research impact
Altogether, this study demonstrated the feasibility of using high-density peptide arrays to identify peptides with repulsive and adhesive properties, offering a new tool that can accelerate discoveries in multiple fields related to cell-surface interactions and biomaterial design:
- The ability to systematically screen thousands of peptides allows for rapid and efficient identification of peptides with specific characteristics.
- The identification of short, functional peptides offers a potentially more cost-effective alternative to using full proteins or synthetic polymers for cell patterning and control.
- Cell-repellent peptides are under-researched, and their identification may offer an alternative to traditional ways of modulating cell interactions.
- The identification of both adhesive and repulsive peptides offers new possibilities for designing biomaterials with precise control over cell behavior, which is particularly relevant for tissue engineering and regenerative medicine applications.
- The discovery that random peptide combinations can have stronger adhesive properties than naturally occurring sequences suggests the potential for developing novel biomaterials that outperform the existing repertoire.
- The discovery of cell-repellent or adhesive motifs on secreted proteins and at specific locations on protein structures provides new insights into protein function and cell-protein interactions.
- The integration of reporter constructs in the screening process opens possibilities for identifying peptides that modulate specific signaling pathways.
- The capacity of the method to screen for peptides that affect cell migration and agglomeration is relevant to cancer research, particularly in studying collective cell migration in carcinomas.
Conclusion
The versatile nature of this method, which enables the evaluation of a vast number of peptides across different scales, ranging from microscopic spots to larger patterns, makes it a valuable resource for several research approaches in the fields of cell biology and bioengineering.
The peptide library used in this study was synthesized via the high-density peptide microarray technology of Axxelera. Do you have projects that could benefit from the use of high-density peptide arrays? Contact us to discuss your specific research needs.
Reference
[1] Sonnentag SJ, Jenne F, Orian-Rousseau V, Nesterov-Mueller A. High-throughput screening for cell binding and repulsion peptides on multifunctionalized surfaces. Commun Biol. 2024 Jul 17;7(1):870. doi: 10.1038/s42003-024-06541-7.