Introduction
The pharmaceutical drug discovery process is a multistep endeavor analogous to a funnel, where vast candidate pools of drugs are gradually narrowed to reduce the number of promising lead compounds that can potentially be used therapeutically.
A critical step in this process is the transition between the large candidate pools generated by high-throughput screening techniques to a practical number of promising leads that can be validated biochemically.
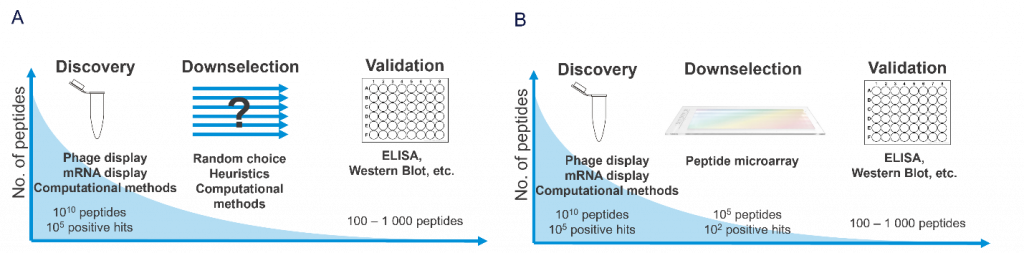
Current screening methods rely on heuristic-driven selection to narrow these vast pools to more manageable numbers (typically dozens or hundreds) for further validation [3]. In this narrowing process, like a bottleneck, many potentially valuable compounds are lost, owing to the inherent limitations of the selection criteria (Figure 1). Overcoming this challenge is crucial for improving the success rates of drug development pipelines. This can be achieved using a quantitative, high-throughput, and data-driven method: peptide microarrays.
Current Screening Methods and their Limitations
The two predominant approaches for assessing large pools of potential drug candidates are display technologies and computational methods:
- mRNA and phage display are powerful in vitro selection techniques that can generate vast combinatorial peptide libraries, typically ranging from 105 to 107 unique sequences. These methods rely on the physical linkage between the phenotype (the displayed peptide or protein) and genotype (encoding nucleic acid), allowing for simultaneous screening and identification of binding molecules [4].
- Computational chemistry approaches, particularly virtual screening, can produce candidate pools of similar magnitude. These in silico methods employ algorithms to screen large virtual libraries of compounds against a target protein structure or pharmacophore model [5].
The critical issue lies in the subsequent narrowing of the resulting vast candidate pools to reasonable numbers of promising leads that can be practically tested by (macro-scale) low-throughput validation assays (ELISA, Western-Blot; Figure 1). The current narrowing methods include random selection, “eyeballing, ” and computational methods (Figure 1A). However, these methods have intrinsic disadvantages: random selection and “eyeballing” are (partially) not data-driven and deterministic and computational methods are limited by the accuracy of their scoring functions and availability of high-quality structural data.
This creates a significant disparity between the size of the initial candidate pool and the number of compounds that can be thoroughly validated (Figure 1). The risk of valuable candidates being discarded owing to the limitations of the screening process is a significant concern in early-stage drug discovery [6].
Peptide Microarrays as a Solution
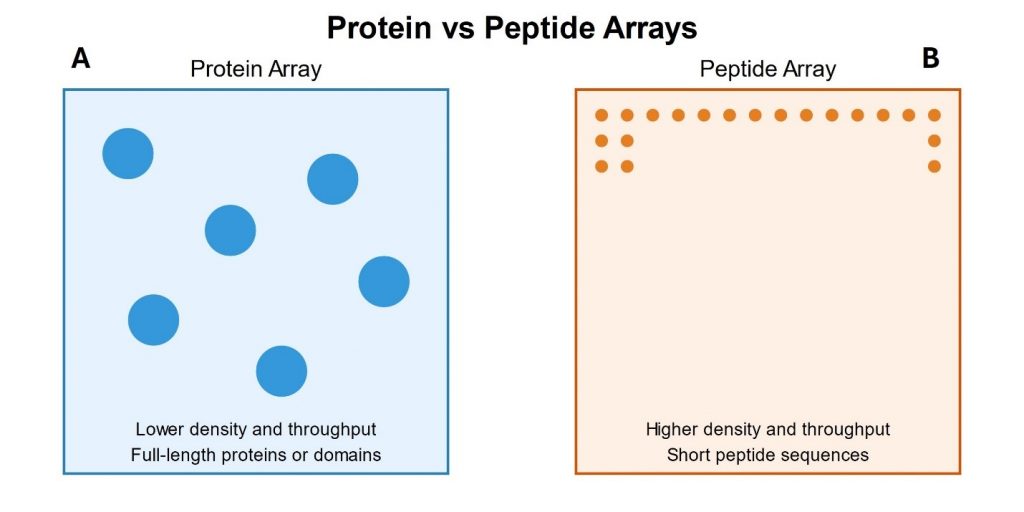
Instead of relying on heuristic-driven selection methods, researchers can use a more quantitative, high-throughput, and data-driven technology: peptide microarrays (Figure 1B. This technique involves the synthesis of hundreds of thousandsof of distinct peptides on a solid support, typically a functionalized glass slide. Thus, all positive hits from a display experiment can be further tested with as little as one array. Each peptide is synthesized at a specific location on the array, allowing parallel analysis of the binding interactions with a target molecule. The speed and precision of peptide microarrays make the screening process faster and more efficient than relying solely on macro-scale assays. This can accelerate the overall drug discovery pipeline and improve the chances of success in the early stages [7].
These microarrays can handle ultra-high-throughput screening and process thousands of candidates in a single experiment. This allows researchers to explore more candidates and increase the likelihood of finding valuable hits [8].The intensity of fluorescence signals serves as a robust proxy for affinity, enabling researchers to quantitatively assess and prioritize potential leads based on their binding affinity, thereby reducing reliance on randomness and the risk of overlooking promising leads [9].
Conclusion
The current bottlenecks in pharmaceutical screening pipelines are clear; vast candidate pools generated by cutting-edge technologies are being narrowed using methods that are inherently limited in their throughput and precision. Peptide microarray technology offers a scalable and quantitative solution to this problem, enabling a data-driven, ultra-high-throughput approach to streamline candidate narrowing in the funnel.
Are you interested in incorporating cutting-edge peptide microarrays into your discovery pipeline? Discover our peptide microarray solutions or contact us directly.
References
[1] Josephson K, Ricardo A, Szostak JW. mRNA display: from basic principles to macrocycle drug discovery. Drug Discov Today. 2014 Apr;19(4):388-99. doi: 10.1016/j.drudis.2013.10.011.
[2] Mimmi S, Maisano D, Quinto I, Iaccino E. Phage Display: An Overview in Context to Drug Discovery. Trends Pharmacol Sci. 2019 Feb;40(2):87-91. doi: 10.1016/j.tips.2018.12.005.
[3] Makley LN, Gestwicki JE. Expanding the number of ‘druggable’ targets: non-enzymes and protein-protein interactions. Chem Biol Drug Des. 2013 Jan;81(1):22-32. doi: 10.1111/cbdd.12066.
[4] Hoogenboom HR. Selecting and screening recombinant antibody libraries. Nat Biotechnol. 2005 Sep;23(9):1105-16. doi: 10.1038/nbt1126.
[5] Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov. 2004 Nov;3(11):935-49. doi: 10.1038/nrd1549.
[6] Macarron R, Banks MN, Bojanic D, Burns DJ, Cirovic DA, Garyantes T, Green DV, Hertzberg RP, Janzen WP, Paslay JW, Schopfer U, Sittampalam GS. Impact of high-throughput screening in biomedical research. Nat Rev Drug Discov. 2011 Mar;10(3):188-95. doi: 10.1038/nrd3368.
[7] Legutki JB, Zhao ZG, Greving M, Woodbury N, Johnston SA, Stafford P. Scalable high-density peptide arrays for comprehensive health monitoring. Nat Commun. 2014 Sep 3;5:4785. doi: 10.1038/ncomms5785.
[8] Loeffler FF, Foertsch TC, Popov R, Mattes DS, Schlageter M, Sedlmayr M, Ridder B, Dang FX, von Bojničić-Kninski C, Weber LK, Fischer A, Greifenstein J, Bykovskaya V, Buliev I, Bischoff FR, Hahn L, Meier MA, Bräse S, Powell AK, Balaban TS, Breitling F, Nesterov-Mueller A. High-flexibility combinatorial peptide synthesis with laser-based transfer of monomers in solid matrix material. Nat Commun. 2016 Jun 14;7:11844. doi: 10.1038/ncomms11844.
[9] Buus S, Rockberg J, Forsström B, Nilsson P, Uhlen M, Schafer-Nielsen C. High-resolution mapping of linear antibody epitopes using ultrahigh-density peptide microarrays. Mol Cell Proteomics. 2012 Dec;11(12):1790-800. doi: 10.1074/mcp.M112.020800.