Exploring RNA-Peptide Interactions Using High-Density Peptide Arrays: A Study Overview
Introduction
Understanding RNA-peptide interactions is a fundamental aspect of molecular biology, particularly when it comes to decoding the origins of genetic translation mechanisms. A recent study, Screening for Primordial RNA–Peptide Interactions Using High-Density Peptide Arrays, conducted in collaboration with Axxelera, takes a closer look at these interactions using advanced screening techniques. The study leverages high-density peptide arrays to explore how primordial peptides interact with RNA, providing key insights into molecular evolution.
This post will walk through the study’s objectives, methods, findings, and its broader implications, demonstrating the utility of high-density peptide arrays in advancing research into the origins of life.
Background
RNA-peptide interactions are central to understanding the evolutionary processes that led to modern protein synthesis and the development of the genetic code. Many evolutionary biologists support the theory of an “RNA/peptide world,” where RNA and short peptides coexisted and co-evolved before the emergence of full-fledged ribosomal translation systems. The stereochemical hypothesis suggests that amino acids may have been selectively paired with their codons early on in the evolution of the genetic code.
However, despite significant computational and bioinformatic progress in identifying RNA-protein complexes, there is still a need for experimental methods that can examine these interactions at a molecular level. The study in question employs high-density peptide arrays to bridge this gap, aiming to explore how individual amino acids contribute to RNA binding.
Methodology
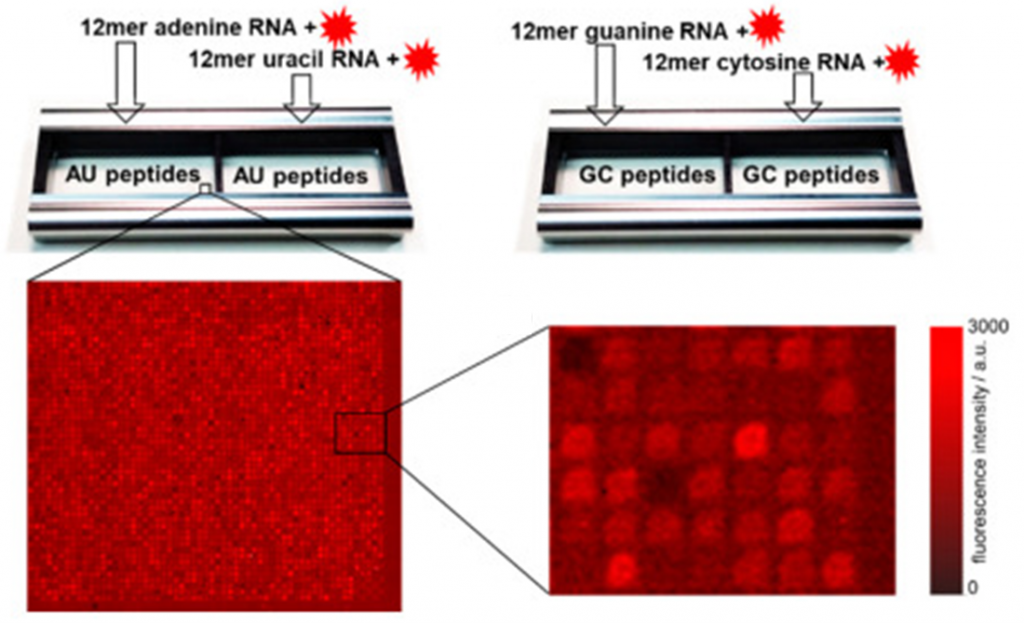
The research employed combinatorial libraries of peptides to test RNA interactions. The peptide libraries were synthesized and arranged on high-density peptide arrays, a technique that allows for the systematic analysis of thousands of peptide variations in parallel. Each peptide library represents a unique set of amino acid combinations, generated according to Watson–Crick base pairing rules to simulate primordial RNA environments.
Axxelera supplied the peptide arrays, which were crucial for the high-throughput nature of the study. The arrays allowed researchers to screen a vast number of potential RNA-peptide interactions simultaneously, ensuring comprehensive coverage of the possible interaction space. The peptides were incubated with fluorescently labeled 12-mer RNA homo-oligonucleotides of adenine, cytosine, guanine, and uracil, under physiological conditions. This approach allowed for the precise measurement of interaction strength through fluorescence intensity, where stronger RNA-peptide binding resulted in higher fluorescence signals.
Findings
One of the study’s key findings was the significant role played by aromatic amino acids—namely phenylalanine, tyrosine, and proline—in strengthening RNA-peptide binding. These results are consistent with earlier studies, which suggest that certain amino acids may have been more likely to participate in primordial RNA-peptide interactions due to their structural properties. For example, phenylalanine and tyrosine, which contain aromatic rings, exhibited strong binding to A-rich RNA sequences.
Interestingly, the charge of the peptides did not significantly influence binding strength, challenging earlier assumptions that charge would be a major factor in RNA-peptide interactions. Instead, the study suggests that molecular weight and hydrophobicity—especially in the context of amino acids like proline—play a more crucial role in determining interaction strength.
Further, the findings supported the theory of combinatorial fusion, which proposes that early genetic codes may have formed through the merging of AU- and GC-rich RNA sequences. This combinatorial fusion is thought to have laid the groundwork for the standard genetic code (SGC), with the study demonstrating how peptides corresponding to different stages of genetic evolution interacted with RNA.
The data also showed a disparity in binding strength between AU- and GC-rich sequences. Peptides exhibited much stronger interactions with GC-rich RNA sequences, possibly indicating that early RNA-peptide interactions took place in environments dominated by G and C bases before expanding to include A and U bases.
Conclusion
The use of high-density peptide arrays provided valuable insights into the molecular dynamics of RNA-peptide interactions, particularly in the context of evolutionary biology. By allowing for the parallel screening of thousands of peptide sequences, the arrays enabled researchers to uncover specific patterns of interaction that would have been difficult to observe using traditional methods. This study highlights the importance of phenylalanine, tyrosine, and proline in early RNA binding, while also supporting the combinatorial fusion model for the origin of the genetic code.
Moreover, this study shows that peptide microarrays can be a useful tool in experimental molecular biology, offering a scalable and efficient method for investigating complex biological phenomena. If you’re looking to explore these innovative tools for your own research, consider contacting us.