The interface between the catalyst and membrane layers in fuel cells is critical for their performance and longevity, thus rendering it a focal point for optimization. Schmidt et al. used high-density peptide microarrays to identify selective peptide binders to Nafion, the most popular membrane material. The peptides identified demonstrated a high affinity for the polymer, showing the potential for enhancing fuel cell membranes in energy technologies. These findings offer insights for the design of advanced materials with implications for energy, environmental, and sensing applications
Background
Polymer electrolyte membrane fuel cells (PEM-FCs) are a promising technology for achieving energy transition goals in various applications, including long-distance transportation and decentralized energy systems. The catalyst-coated membrane (CCM) is at the heart of PEM-FCs, where energy-converting reactions occur. Improving the interface between the catalyst layer and membrane is crucial for enhancing fuel cell performance and durability.
Nafion, a perfluorinated sulfonic acid (PFSA) polymer, is the most widely used membrane material in state-of-the-art fuel cells. Recent research has focused on structuring ion channels at the molecular level to improve mass transfer in the membrane electrode assembly. However, to exploit this approach, new functional molecules that can selectively bind to the Nafion ionomer are required.
The recent study by Schmid et al [1] proposed using peptides as promising templates for this task, as they have previously shown promise in the field of energy storage and conversion materials. The researchers developed a method to screen for peptide binders to Nafion using ultra-high density peptide arrays.
Methodology
- Initially, the researchers used ultra-high-density peptide arrays to screen for optimal selective peptide binders for Nafion dissolved in solution. The initial library contained 56,014 randomly selected 6-mer peptides that were synthesized on a conventional microscope slide. The peptide chip was incubated with a diluted Nafion dispersion and scanned using a confocal fluorescent scanner to detect the ionomer molecules on the peptide spots.
- Subsequently, the authors selected the best performing peptide (WIWHCW) and created a substitution library to determine the importance of each amino acid in the sequence. They further conducted dilution experiments to determine the dissociation constants of the peptide binders.
- Lastly, the authors used Nuclear Magnetic Resonance (NMR) spectroscopy to validate Nafion-peptide binding. Additionally, they studied the adhesion of the selected peptide on Nafion membranes using fluorescently labeled peptides.
Key findings
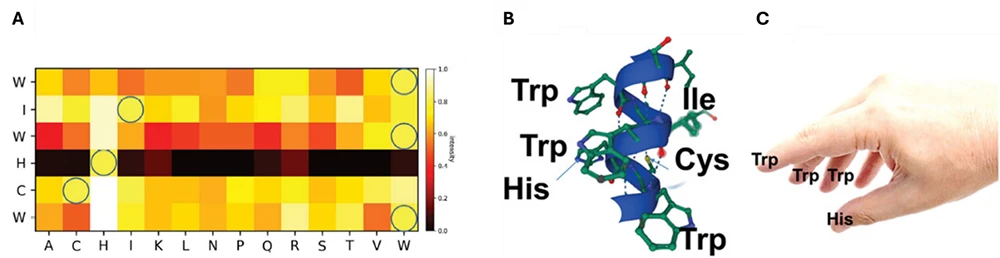
- The peptide WIWHCW was identified as a selective binder for Nafion, forming a helix structure with a binding pocket created by histidine (H) and tryptophan (W) residues (Figure 1).
- Substitution analysis revealed that histidine plays a significant role in binding to the ionomer, whereas replacing cysteine (C) and isoleucine (I) did not significantly affect the interaction (Figure 1A).
- Adding additional histidine to the peptide sequence in the presence of three or two tryptophan residues increased the binding strength between the peptide and ionomer. This insight provides valuable information for designing potentially stronger binders in the future.
- The dissociation constant of WIWHCW to Nafion was measured at approximately 140 µM, which is weaker than antibody-antigen interactions but within the range of most protein-protein interactions in cells.
- NMR measurements confirmed ionomer binding via significant line broadening in the aromatic and methyl regions of the peptide spectrum.

Research impact
- This study demonstrates the novel use of ultra-high-density peptide arrays for screening selective peptide binders for technical polymers, such as Nafion. The method allows for high-speed, parallel screening and validation of results.
- The identified peptide binders have potential applications in the design of novel virus-templated materials and structuring ion channels in fuel cell membranes.
- The overall approach can be used to improve the performance and efficiency of PEM-FCs generally, contributing to the advancement of energy technologies.
Conclusion
The researchers successfully developed a method to screen for selective peptide binders to Nafion using ultra-high-density peptide arrays. They identified a family of selective binders and measured their dissociation constants, gained structural insight into the specificity of the binding mechanism, and laid out a promising technology to improve fuel cell technology.
The high-density peptide microarray used in this study was provided by Axxelera. Up to 200,000 peptides can be analyzed per experiment with Axxelera’s technology, enabling the execution of even larger-scale experiments in a time- and cost-efficient manner. Contact us to learn how we can assist with your projects.
References
[1] Schmidt D, Gartner P, Berezkin I, Rudat J, Bilger M, Grünert T, Zimmerer N, Quarz P, Scharfer P, Brückel J, Jung AP, Sing P, Pooja P, Meier B, Stahlberger M, Schabel W, Bräse S, Lanza G, Nesterov-Mueller A. Selective Peptide Binders to the Perfluorinated Sulfonic Acid Ionomer Nafion. Adv Funct Mater. 2023, 34, 2214932. https://doi.org/10.1002/adfm.202214932